Enzyme may help bacteria become craftier and more dangerous

Virginia Tech researchers have discovered a new role for an enzyme that is well-known for fueling chemical reactions in bacterial cells.
The enzyme, an ATPase known as PilB, is part of a biological chain reaction that allows bacteria to respond to changes in the environment.
The finding, published in Scientific Reports, will help scientists better understand how dangerous bacteria, such as pathogenic E. coli and Pseudomonas, create biofilms to colonize the human body and medical implants, including pacemakers. Bacteria in biofilms can make infections hard to treat, resulting in serious medical problems.
It was previously known that PilB is used to break down adenosine triphosphate, or ATP, a small energy-storing molecule. But now, researchers know that it can help bacteria process information as well, such as how to respond to changes in their environment.
“There is a hidden way for cells to do things that we didn’t know before,” said Zhaomin Yang, an associate professor of microbiology in the College of Science at Virginia Tech, Fralin Life Science Institute affiliate, and lead author on the paper. “This is an alternative way signals can be processed, much like cell phones process and transmit signals differently from landlines.”
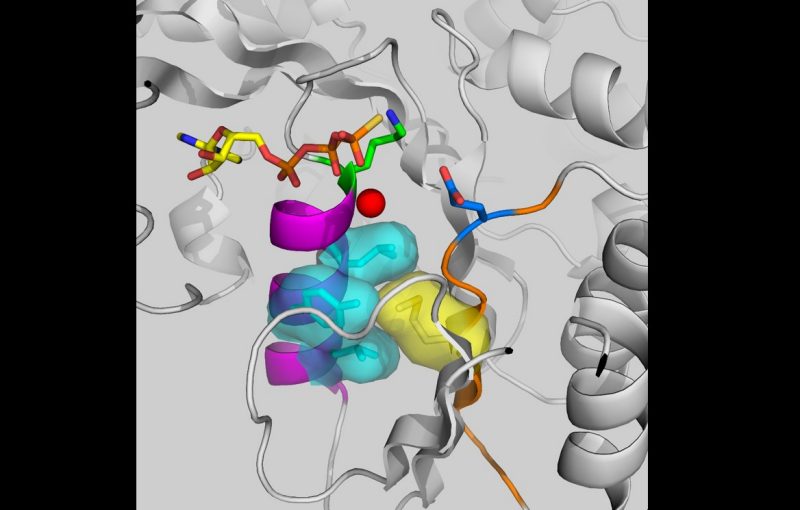
Yang and his colleagues made the discovery while studying how myxobacteria form biofilms. These bacteria, which are commonly found in the environment, colonize by secreting a sticky, glue-like substance that allows them to spread on moist solid surfaces to look for nutrients. Likewise, pathogenic bacteria form biofilms in nutrient-rich environments, such as the human body. In both of these situations, bacteria use environmental signals – the presence or absence of nutrients – to determine whether to produce this matrix material for biofilms.
In this study, the myxobacterium Myxococcus xanthus was found to regulate biofilm formation and bacterial movement through the PilB ATPase enzyme, which was already known to be essential for bacterial movement on surfaces.
“The surprising finding here that an ATPase can function as a signaling protein will likely impact biological and biomedical research in profound ways,” Yang said. “There are typically dozens if not hundreds of ATPases in a given organism. It is exciting to envision the prospect of these enzymes forming the wiring of signal processing networks in a cell, from bacteria to animals.”
Scientists have known that ATPase enzymes harvest energy from ATP in order to fuel such processes as movement of a cell or organisms, explained Yang, but prior to this discovery, little was known about whether they could directly function as signaling proteins in biological systems. Other proteins, called G proteins, have been the ones known to play this role.
“Before now, ATPases were thought to provide energy for various reactions in the cell,” Yang said. “This could be the beginning of uncovering many new ways cells manage signals, significantly broadening the spectrum of how cells process and respond to signals.”
In addition to Yang, other members of the research team include Wesley Black, a postdoctoral associate in microbiology; Lingling Wang, a visiting scientist from China; and Birgit Scharf and Florian Schubot, both associate professors of biological sciences in the College of Science and affiliates of the Fralin Life Science Institute.
This research was supported in part by the National Institutes of Health, the American Heart Association, and the National Science Foundation.
Written by Cassandra Hockman






.jpg.transform/m-medium/image.jpg)