Researchers discover new mechanism of antibiotic resistance in leprosy and tuberculosis
Researchers

A Virginia Tech research team in collaboration with researchers from the University of Missouri-Columbia have discovered a mechanism responsible for antibiotic resistance in the bacteria that cause tuberculosis and leprosy.
Knowledge of this mechanism will ultimately allow researchers to design more effective drugs to treat these diseases.
These findings were recently published in the journal Biochemistry.
Rifampicin, a popular antibiotic used to treat tuberculosis and leprosy, works by preventing bacteria from growing. However, the drug is becoming less effective as Mycobacterium tuberculosis, the bacteria that cause tuberculosis, and Mycobacterium leprae, the bacteria that cause leprosy, develop resistance.
This resistance occurs in part because the bacteria have a certain enzyme, called rifampicin monooxygenase, that inactivates rifampicin by chemically modifying its structure.
Pablo Sobrado, a professor of biochemistry in the College of Agriculture and Life Sciences, and his team worked in collaboration with biochemist John Tanner and postdoctoral associate Li-Kai Liu at the University of Missouri-Columbia to discover that rifampicin monooxygenase converts rifampicin from a cyclic or ring form to a linear form. Once rifampicin is in this linear form, it is no longer effective in killing the bacteria.
Chemical reaction
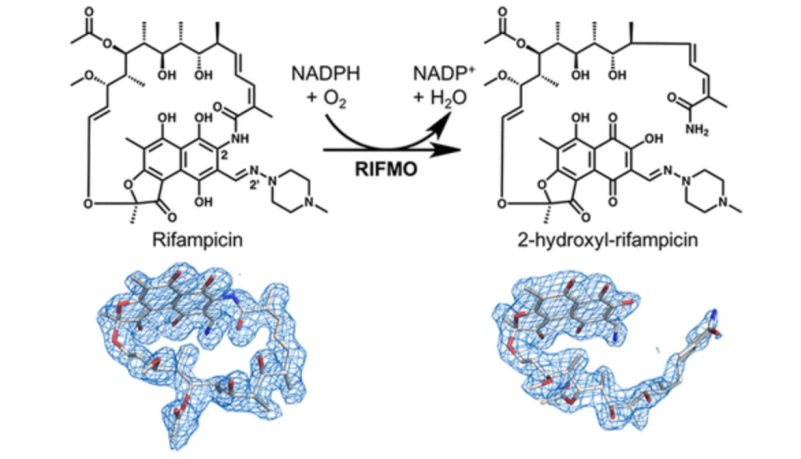
“This discovery is exciting because it is all-important for drug design,” said Sobrado, an affiliate of the Fralin Life Science Institute and the Virginia Tech Center for Drug Discovery. “It can help researchers design new rifampicin analogs that are more effective because they are not susceptible to inactivation by this bacterial enzyme.”
Members of Sobrado’s team who helped identify the mechanism include Yumin Dai, currently a senior research associate in the Department of Chemistry in the College of Science, and visiting graduate student Heba Abdelwahab of Damietta, Egypt. In a previous study, Abdelwahab described the atomic and molecular structure of rifampicin monooxygenase using X-ray crystallography.
In this recent study, funded with a National Science Foundation grant, Sobrado’s team used X-ray crystallography, NMR analysis, and mass spectroscopy to identify the chemical modification of rifampicin by rifampicin monoxygenase. Before this finding, researchers knew that rifampicin monoxygenase inactivated rifampicin, but they did not understand how.
“Antibiotic resistance is an evolving issue that greatly impacts the public health,” said Abdelwahab. “Our studies have shown how this enzyme deactivates rifampicin. We now have a blueprint to inhibit this enzyme.”
Dai, who conducted this research as a postdoctoral associate in Sobrado’s lab, added, “based on understanding this mechanism, the next step in research may focus on developing various enzyme inhibitors in order to treat the antibiotic resistance, such as structurally simplified rifampicin mimics, that would act as competitive inhibitors.”
Drug researchers can also begin to design improved forms of rifampicin via synthetic modification that are not susceptible to chemical modification by the bacterial enzyme that targets it.
While treatable, tuberculosis and leprosy are diseases that pose a threat to children, the elderly, people in developing countries without access to adequate health care, and others with compromised immune systems. According to the Centers for Disease Control and Prevention, more than 10 million people worldwide were sick with tuberculosis in 2016. The rates and prevalence of leprosy are much less; globally, less than one case per 10,000 people is reported each year, and 96 percent of these cases are from developing countries.
Written by Kristin Rose





.jpg.transform/m-medium/image.jpg)

